marinemicrobes
Marine Microbes Data Tools
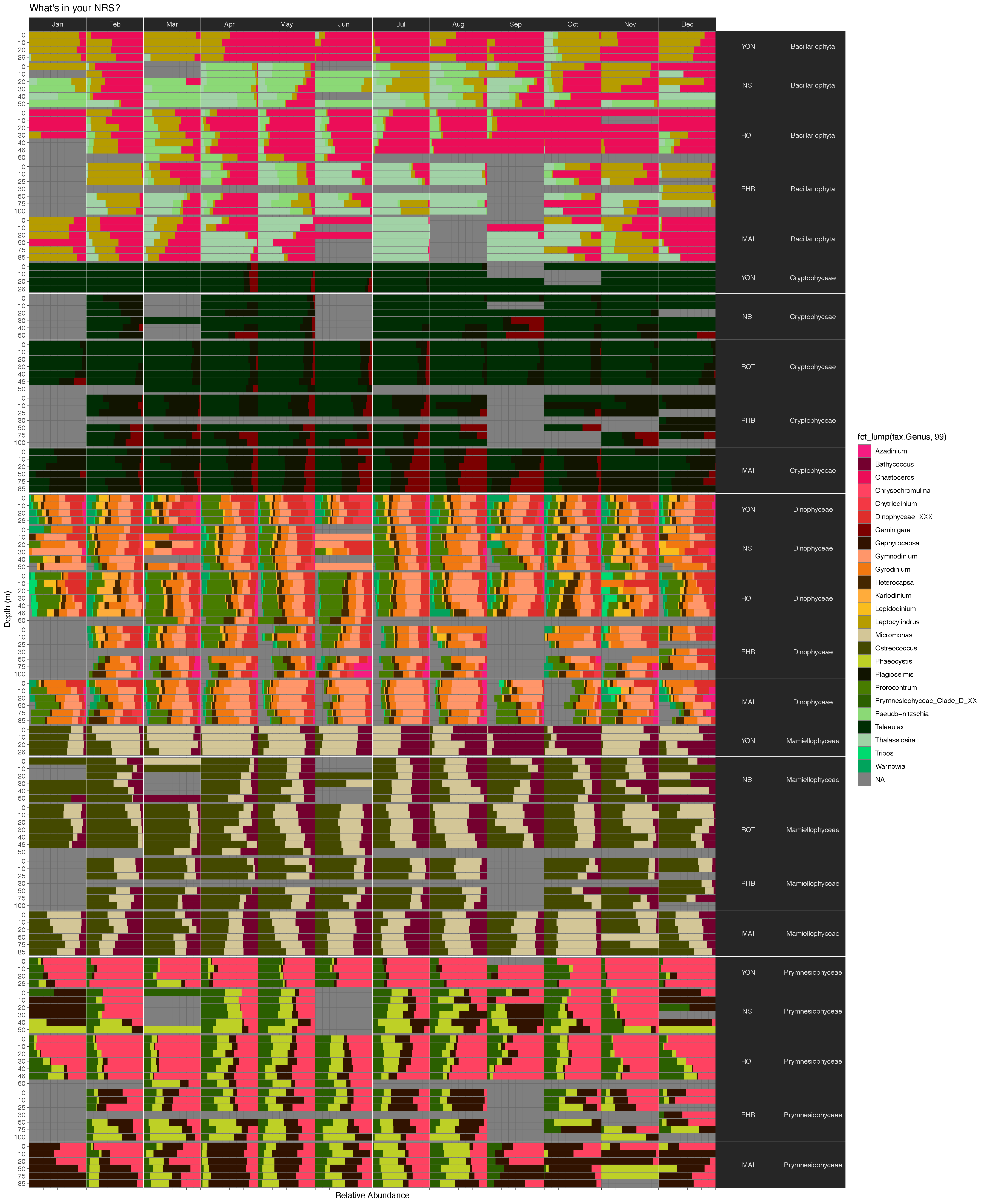
Amplicon analysis workflow
- Raw data download
- Amplicon Sequence Pipeline
- Assembling sequence variant tables
- Taxonomic assignment using DADA2
- Comparison between methods
- Ecological Statistics
- Ordination, visualisation and presentation
- Community Thermal Index
- Boosted Regression Tree Modelling
Metagenome analysis workflow
- Assembly
- Co-assembly
- Binning and bin-refining
- Metagenome Assembled Genomes (MAGs)
- Annotation
- Metatranscriptome analyses
Additional routines
- Spatial mapping SST, Chl
- PAR
- Cyano Reference Database
- HPC tools
- Interactive Maps
- Flow cytometry
Publications
Contributors
Dr Deepa Varkey, Dr Amaranta Focardi, Dr Mark V. Brown, Dr Anna Bramucci, Dr Beth Signal, Dr Tim Kahlke, Geraldine Sullivan (Informatics officer), and James O’Brien (PhD Candidate, Expeditioner and Research Associate)
Overview
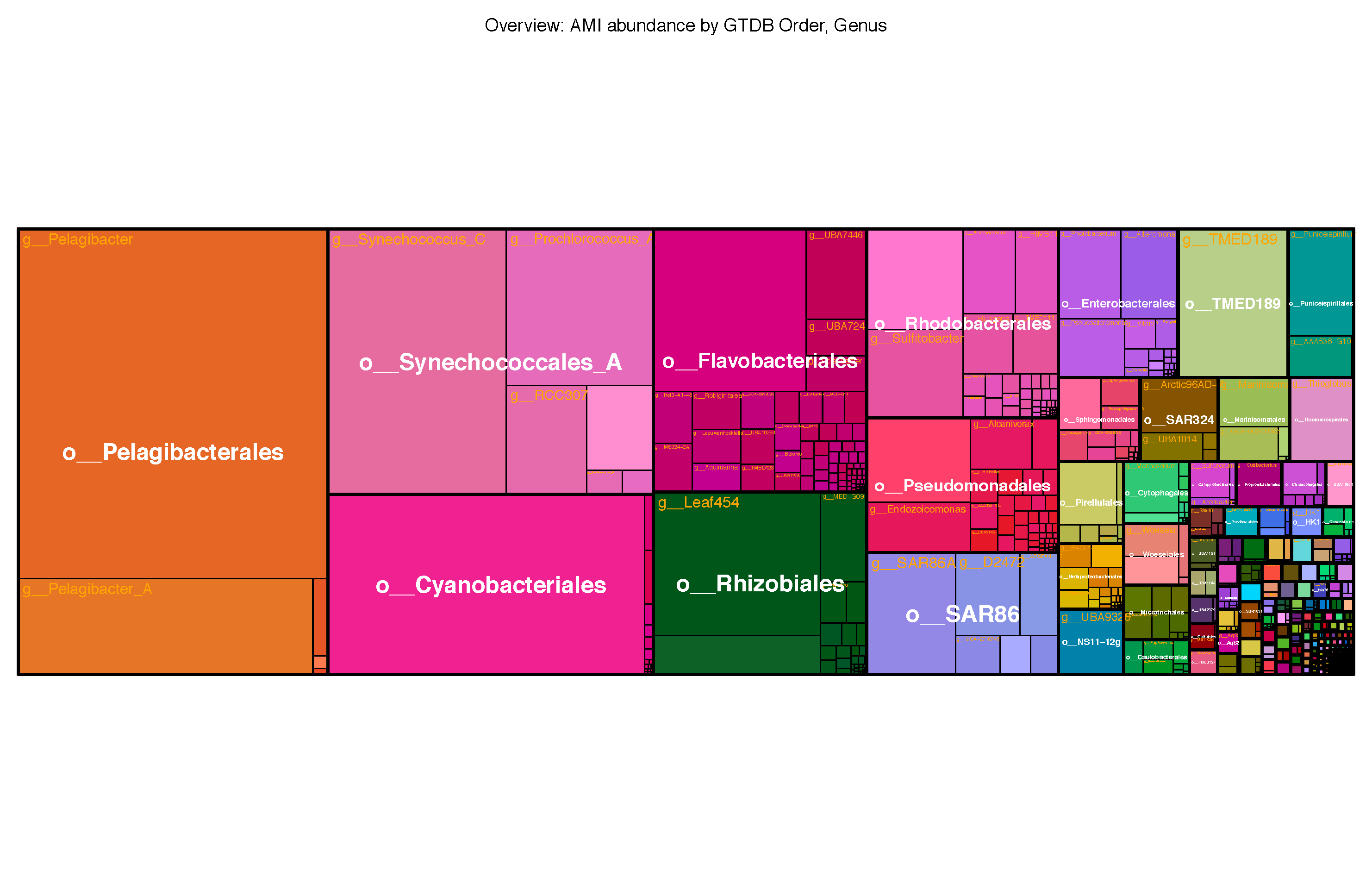
Contact
Martin Ostrowski. Ocean Microbiology Group, C3 Institute, University of Technology, Sydney email: Martin Ostrowski
Uniques data was processed from The Australian Microbiome raw data by Matt Smith, Andrew Bissett & Mark Brown
Acknowlegement
We acknowledge the contribution of the AMMBI Project and Marine Microbes consortium in the generation of data used in this publication. The Marine Microbes project was supported by funding from Bioplatforms Australia through the Australian Government National Collaborative Research Infrastructure Strategy (NCRIS) in partnership with the Australian research community.
References
- The Australian Microbiome Data Portal
- The Genome Tree Database GTDB
- The Protist Reference Database 2 PR2